Complex biogeochemical networks that include many biotic and abiotic processes can be limited by different substrates under different conditions during their temporal evolution.

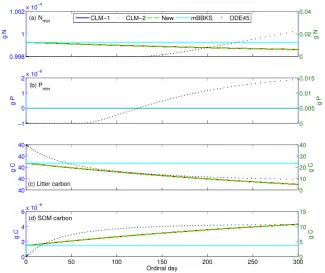
Existing methods are either cumbersome to implement or unable to address this challenge. However, these processes are critical to model important biogeochemistry-climate feedbacks, such as nitrogen and phosphorus regulation of terrestrial carbon exchanges. Therefore, Jinyun Tang and Bill Riley (LBNL) developed a stoichiometry based flux-limiter approach to modify the Euler forward ordinary differential equation integrator and consistently implemented the substrate limitation for biogeochemical networks of arbitrary complexity. When applied to a Century-like biogeochemical network involving carbon, nitrogen, and phosphorus, Tang and Riley showed that this new approach gave consistent predictions compared to the ad hoc approaches used in CLM and the ACME Land Model (ALM), while the existing single-limiter approach (mBKKS, Figure 4) and clipping approach (ODE45, Figure 4) predicted incorrect decomposition dynamics (Figure 4). Moreover, their approach is immune to the “ordering problem”, which CLM and ALM suffer from. We are integrating this approach in the Equilibrium Chemistry Approximation-based version of ALMv1 to represent carbon, nitrogen, and phosphorus coupled biogeochemistry. This approach will improve simulations of high-latitude, temperate, and tropical coupled plant and soil biogeochemistry and the resulting C-climate feedbacks.
Reference:
Tang, J. Y. and Riley, W. J.: Technical Note: A generic law-of-the-minimum flux limiter for simulating substrate limitation in biogeochemical models, Biogeosciences, 13, 723-735, DOI:10.5194/bg-13-723-2016, 2016.